Ali Toghani
Papers & Publications
Schimek, N., Wood, T., Beck, D.A.C., Toghani, A., McKenna, M., Nance, E.* "High fidelity predictions of diffusion in the brain microenvironment."Biophysical Journal, Vol. 123, Issue 22, 2024, pp. 3935–3950.https://doi.org/10.1016/j.bpj.2024.10.005
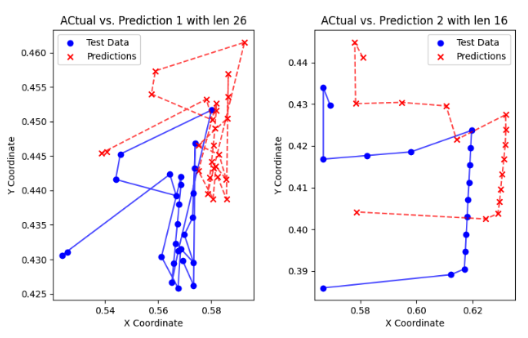
Deep Learning with Multiple Particle Tracking (MPT)
Our research project in Dr. Nance's Lab at the University of Washington focuses on applying advanced deep learning techniques to Multiple Particle Tracking (MPT) data. The primary goal is to enhance the accuracy of predicting nanoparticle trajectories, which is crucial for various scientific and industrial applications. By developing and implementing sophisticated neural network models, we aim to uncover intricate patterns and interactions within the MPT data that traditional methods may miss. This innovative approach not only advances our understanding of particle behavior but also contributes to the broader field of data analysis in nanotechnology. We are also creating an open-source Python package to facilitate MPT data analysis, making these cutting-edge techniques accessible to researchers worldwide. This project is supported by the Washington Research Foundation Fellowship for 2023-2024, highlighting its potential to make significant contributions to the field.
Skills: Python, Pytorch, Machine Learning, Deep Learning, LSTM, Transformers
Github Link: https://github.com/BeckResearchLab/diff_transformers/tree/main/project
Awards: 2023 - 2024 Washington Research Foundation (WRF) Fellowship
Presentations: SciPy 2024 Poster Presentation, Mary Gates Symposium
Data Visualization Notebook
This Visualization Notebook was developed to enhance the efficiency of creating visualizations in Dr. Nance's lab. It provides pre-written code templates for various types of visualizations, allowing researchers to modify and adapt them easily by changing the input file and adjusting parameters. This approach eliminates the need to write code from scratch, saving time and effort. The notebook offers flexibility with options to customize the appearance and functionality of visualizations, making it suitable for a range of data visualization needs. Its intuitive interface ensures easy navigation and accessibility, even for those with limited coding experience. By automating much of the visualization process, the notebook ensures consistency and accuracy in data representation. This tool is invaluable for the lab, supporting effective data analysis and the clear communication of research findings, ultimately advancing scientific understanding through innovative visualization techniques.
Skills: Python, Matplotlib, Cython, Pandas
Github Link: https://github.com/AliToghani82/Visualization_Notebook
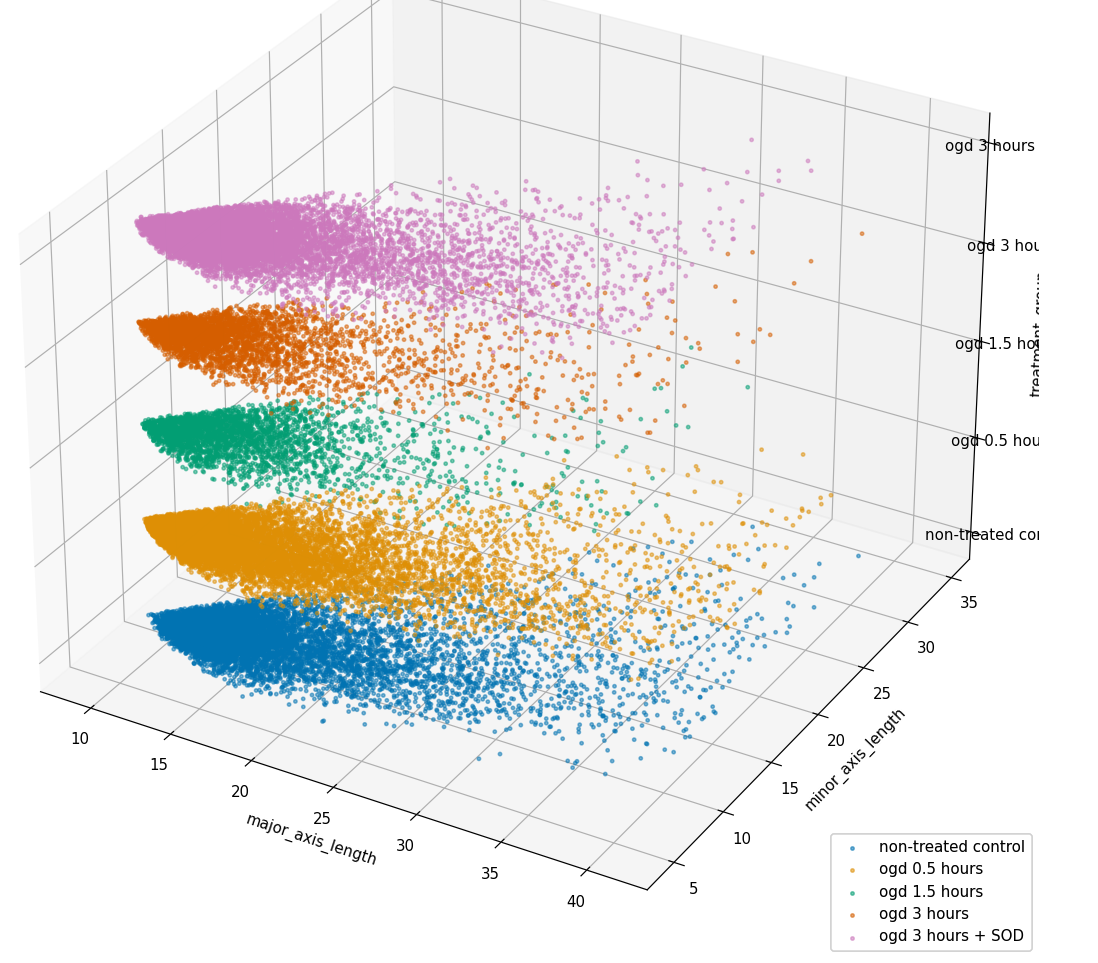
Understanding and evaluating shapes of microglia using geomStat
Integrated the GeomStat package into the data processing pipeline to analyze and evaluate microglia shapes, enhancing the understanding of their structural characteristics
Skills: Python, GeoMstats
Pipeline optimization using Cython
Applied and expanded my knowledge of C++ and Cython to optimize a fluorescent image processing pipeline for ex-vivo brain slices. I also Enhanced science communication skills using Html and JavaScript to present my research progress during meetings
Skills: Python, C++, Cython, Pandas, Javascript, HTML, CSS
Github Link: https://github.com/AliToghani82/Visualization_Notebook